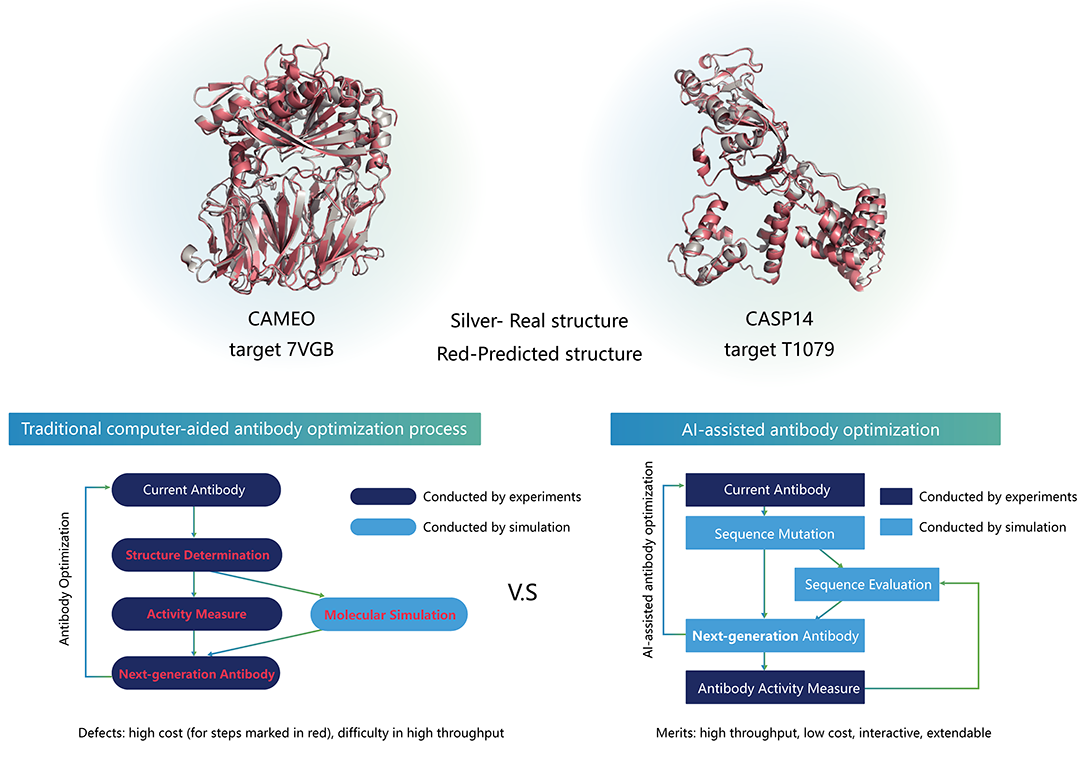
No. 1 in monthly CAMEO contest, protein structure prediction toolkit surpassing AlphaFold 2
2023-01-17
Collaborators: Huawei, Peking University, Shenzhen Bay Laboratory
Built and released the largest known (to release date) open-source protein multiple sequence alignment (MSA) dataset
Built the world's largest open-source protein MSA dataset with the latest reference dataset and the widest coverage by the release date. This protein MSA dataset is searched and aligned with the “gold standard” search method, on ~50 million protein sequences, which is useful in solving problems of highly variable sequences and orphan sequences in protein research.
Developed protein structure prediction toolkit based on the Ascend-MindSpore domestic AI framework, which shows comparable accuracy to AlphaFold2 and outperforms the SOTA model in many aspects
Provide protein structure inference tool based on AlphaFold 2 algorithm, and further developed and benchmarked model training protocol as well as independent checkpoint on the Ascend-MindSpore domestic AI framework. A single-step iteration takes 12 seconds instead of 20 seconds by AlphaFold 2 under mixed precision, reducing time cost by over 60%.
Top in Continuous Automated Model Evaluation (CAMEO) for weeks
Coming out on the top in CAMEO, a community-wide evaluation which continuously evaluates the accuracy and reliability of protein structure prediction servers, through continuous improvement in algorithm, scale, software/hardware configuration. filling the gap in domestic AI software/hardware frameworks in the field of protein structure prediction.